Information architecture
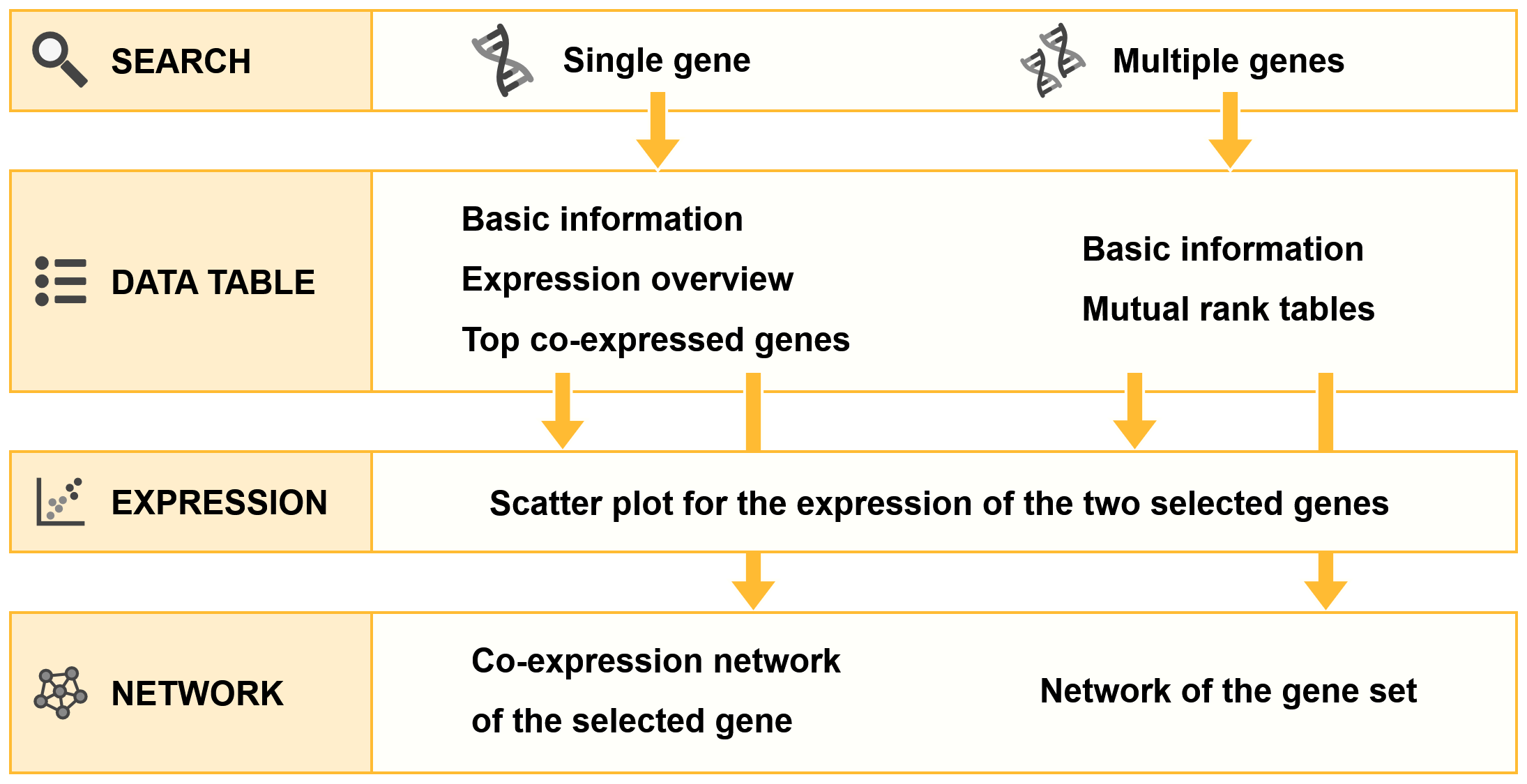
Construction of the co-expression network
1. Raw RNA-Seq read files were trimmed and mapped to the plant genome.
2. Genes with CPM (count per million) ≥ 1 in 20 or more samples were kept for downstream computation.
3. Gene counts were normalized by TPM (transcripts per million) and log2 transformed.
4. PCC (Pearson correlation coefficient) was computed 1000 times for each gene pair, each time with 80% randomly selected samples. The results were averaged.
5. PCC matrix was transformed to MR (mutual rank) matrix. MR was used to rank the co-expression.
Data Download
Raw data
Gene count (tab-separated values)
Co-expression data
These files contain gene pairs with mutual ranks < 1000. Data are stored as tab-separated values.
| Study ID |
| Sample ID |
|---|
| Tissue |
| Stage |
| Treatment |
NODE SIZES small ... high mutual rank to center gene
