
Biomedical Sciences Resources
General Description
The Heritage College Histology Core Facility provides use of the following tissue preparation equipment:
- Leica ASP300S Tissue Processor
- HistoCore Tissue Embedder Arcadia C
- Leica RM2235 Low Profile Microtome
- Leica RM2135 High Profile Microtome
- Leica Autostainer XL
- Cryostat Leica CM1950
The following equipment aids with slide capture:
- LMD Leica 6000
- Motic Slide Scanner
Histology Core Equipment
Tissue Processor Leica ASP300S
The Leica ASP300S is an innovative, smart processor for paraffin infiltration of tissue. The equipment can process a large number of tissue samples simultaneously, including formalin, alcohol, xylene and paraffin. At the end of the process, the samples are ready for paraffin block preparation.

HistoCore Tissue Embedder Arcadia C
The HistoCore Arcadia is a programmable tissue embedding station. Once the infiltrations have been completed in the tissue processor, the samples are ready for block preparation.

Microtome Leica RM2235
The Leica RM2235 is a low-profile microtome that cuts paraffin-embedded tissues for slide preparation. The thickness of the sections can be 3 to 15 microns.

Leica Autostainer XL
The Leica Autostainer XL is a hematoxylin and eosin stainer.

Cryostat Leica CM1950
This instrument is a high precision microtome, which prepares tissue sections in frozen tissue samples usually embedded in OCT.
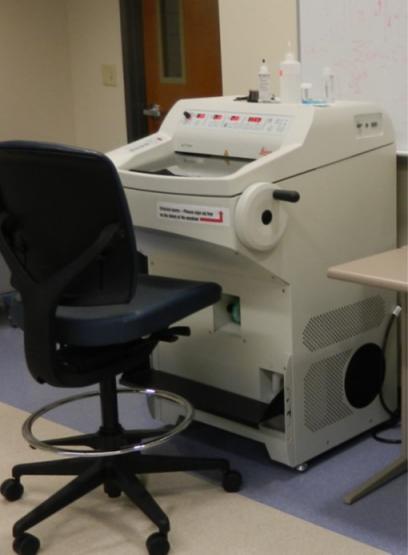
LMD Leica 6000
The LMD Leica 6000 conducts laser capture microdissection and is a standard technique for sample isolation from heterogenous samples. It is commonly used for genomics, transcriptomics, diagnostics and proteomics applications.

Motic Slide Scanner
This slide scanner prepares digital images from slides. The images can be seen on a computer using the associated software.

Details on Using Facility
Fees
| Process | Equipment Required | Rate | Unit |
|---|---|---|---|
|
Embedding Only
|
HistoCore Arcadia H | $ 2.00 | Per cassette |
| |
HistoCore Arcadia C | ||
|
Tissue Processing and Embedding
|
Leica ASP300 | $ 5.00 | Per cassette |
| |
Leica ASP300S | ||
| |
HistoCore Arcadia H | ||
| |
HistoCore Arcadia C | ||
|
Unstained Slide
|
Leica RM2235 | $ 2.00 | Per slide |
| |
Leica RM2135 | ||
|
H&E Staining
|
Leica Autostainer XL | $ 5.00 | Per slide |
| |
Leica RM2235 | ||
| |
Leica RM2135 | ||
|
PAS Staining
|
Leica Autostainer XL | $ 9.00 | Per slide |
| |
Leica RM2235 | ||
| |
Leica RM2135 | ||
|
Motic Slide Scans
|
Digital Scanner | $ 5.00 | Up to 6 slides |
|
Cryostat Slides
|
Leica CM1950 Cryostat | $ 10.00 | Per hour |
|
LMD Slides
|
Leica LMD 6000 | $ 5.00 | Per slide |
Services Provided
The faculty and staff of the Histology Core Facility are highly skilled in the following services:
- Tissue processing
- Paraffin block preparation
- Sectioning
- Hematoxylin & eosin stain
- Other histochemical staining (i.e., PAS, Trichrome)
- Cryostat sections training
Histology Core Summary for Grant Proposals
Requesting Lab Services
- Download and complete the Histology Core Facility Request Form .
- Email the completed form to Noriko Kantake, PhD
to schedule services. Please include the following in your email:
- Brief description of the service you are requesting
- Upcoming deadlines for your request (if applicable)
- After receiving a scheduled time for your requested service, drop off your samples and holders in the Academic and Research Center, Room 240, on the “incoming shelf.” A completed services form (including a billing account number) will need to be placed with the samples. Samples will not be processed without a completed form.
- You will receive an email when the services are complete.
Location
Academic and Research Center, Room 240
Access and Operating Guidelines
The facility is available to the entire Ohio University research community. All users must complete a request for services form, which will be reviewed by the lab technician. Upon approval, the user will be contacted by the lab technician to schedule one-on-one training using their specimens. All users must complete training with the technician before being authorized to use the equipment. Equipment use is by appointment only.
User Responsibilities
Users must provide samples that are ready for preparation. Each user is also responsible for supplying details of any hazards (e.g., infectious organisms, blood-borne pathogens, toxic chemicals) associated with the preparation in accordance to Ohio University Laboratory & Radiation Safety regulations (740-593-1666).
Publication and Acknowledgements - Very Important
To accurately reflect the assistance this resource played in your research and to help ensure its continued funding, please include the following statement in all publications: "We acknowledge use of the Ohio University Heritage College Histology Core."
Acknowledgement and Co-authorship Policies
Co-authorship on publications for staff of the Histology Core may be considered appropriate when they have provided one or more of the following:
- Significant intellectual contribution to the design of the published experiments
- Substantial practical contribution to the generation, analysis and/or interpretation of experimental data
- Indispensable technical support that contributed intellectually or scientifically to the advancement of the work
Staff who have made an intellectual or technical contribution not justifying authorship as defined above should be given appropriate acknowledgement in any resulting publications. This provides evidence to demonstrate our involvement in the research of multiple laboratories, which will help us retain dedicated support staff, maintain the equipment and procure new equipment in the future.
Problem Management
Lab Coordinator
Noriko Kantake, PhD
740.593.2352 or 740.593.2816
278A Academic Research Center
Director
Thomas Rosol, DVM, PhD, MBA
740.593.2405 (office)
Irvine Hall
