Current Research Projects

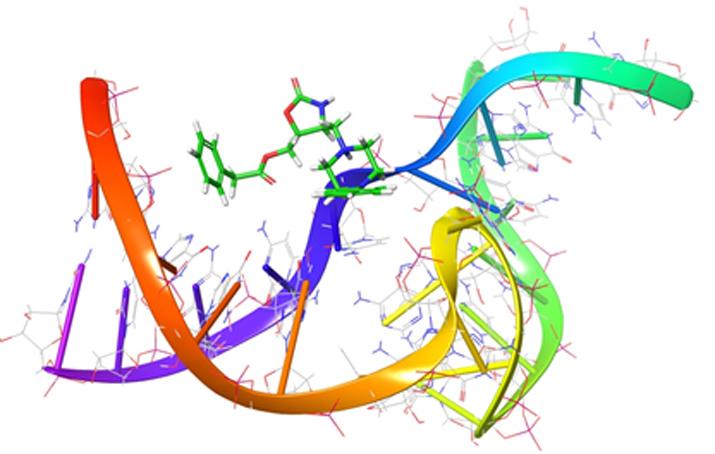
Riboswitch-targeted Drug Discovery
Bacterial drug resistance is a significant global health threat that has turned once treatable bacterial infections (including Gram-positive infections) into deadly illnesses. To overcome this serious threat to human health, completely novel antibiotics are needed. Noncoding ribonucleic acids regulate a wide array of important cellular processes and represent a novel set of targets for developing the next generation of therapeutic agents to treat infectious diseases. Our lab is currently investigating strategies for targeting and disrupting the T-box riboswitch. This molecular switch is a noncoding RNA that regulates the expression of essential genes in many Gram-positive bacteria (including pathogenic examples), thus making it a unique target for novel antibacterial agents. A key component of the T-box riboswitch regulatory mechanism involves tRNA binding to the highly conserved antiterminator RNA element--an excellent target for drug discovery. We are combining structural biology, molecular biology, bioanalytical chemistry and molecular modeling strategies to discover novel compounds which target this RNA specifically and may ultimately be developed in to new antibacterial treatments.
RNA Thermal Stability and Drug Discovery
The thermal stability of an RNA secondary and tertiary structure is both a physical characteristic and also a potential target for drug discovery. This is particularly the case with RNA thermoregulators in bacteria that regulate gene expression via the thermal denaturation of unique RNA elements. Our lab is currently investigating environmental factors (temperature, cations, small molecules and other biomolecules) that affect RNA thermal stability and the extent to which ligand structure-specific interactions can be designed to alter this stability selectively.
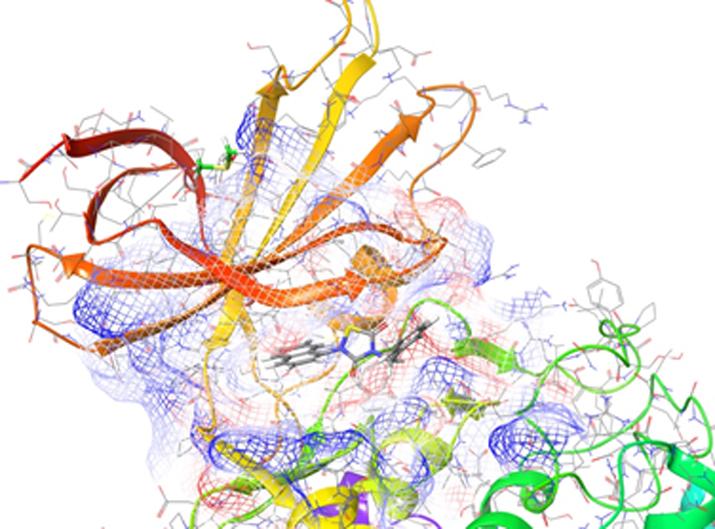
Computational Drug Discovery
We use molecular dynamics to model structurally complex RNA motifs and identify unique, druggable conformers. We have applied this approach to studying unique RNA motifs in bacteria and viruses, including SARS-CoV-2, the virus causing COVID-19. In addition to the computational research in the RNA drug discovery projects, we are also investigating ligand-protein interactions computationally to design novel compounds to treat cancer and diabetes. We are researching the structure-activity relationship of small molecules binding to key regulatory receptors. We utilize a combination of bioinformatic, molecular dynamics and molecular modeling techniques to identify key recognition features in the ligand-receptor interactio n.
